SP B4
Network analysis of HDAC-controlled microRNAs in neuroblastoma
The importance of epigenetics in cancer biology has become evident in recent years. In neuroblastoma, a childhood cancer derived from the developing sympathetic nervous system, high-level amplification of the MYCN oncogene correlates with poor overall survival in patients. An imbalance between histone deacetylase (HDAC) and histone acetyltransferase activity can create changes in the physiological patterns and levels of protein acetylation. We and others have shown that inhibiting HDAC and MYCN activity triggers strong antitumoral effects in cellular and animal models of neuroblastoma, and several microRNAs have been identified that play important roles in neuroblastoma cell proliferation, migration, invasion and metastasis. Dissecting the microRNA network controlled by HDAC and MYCN in neuroblastoma cells has the power to identify both microRNAs and their protein-encoding RNA targets that are critical vulnerable nodes for pharmaceutical targeting in the neuroblastoma transcriptome. Biomarkers predicting the response to HDAC inhibitor (HDACi) treatment in neuroblastoma patients have not yet been identified, but are necessary for patient stratification before this type of targeted therapy can be integrated into treatment schedules. The overall goal of SP B4 is to identify biomarker networks in a systems biology approach and develop strategies for predicting the response to HDAC inhibitor treatment in NB patients and validate the outcome experimentally.
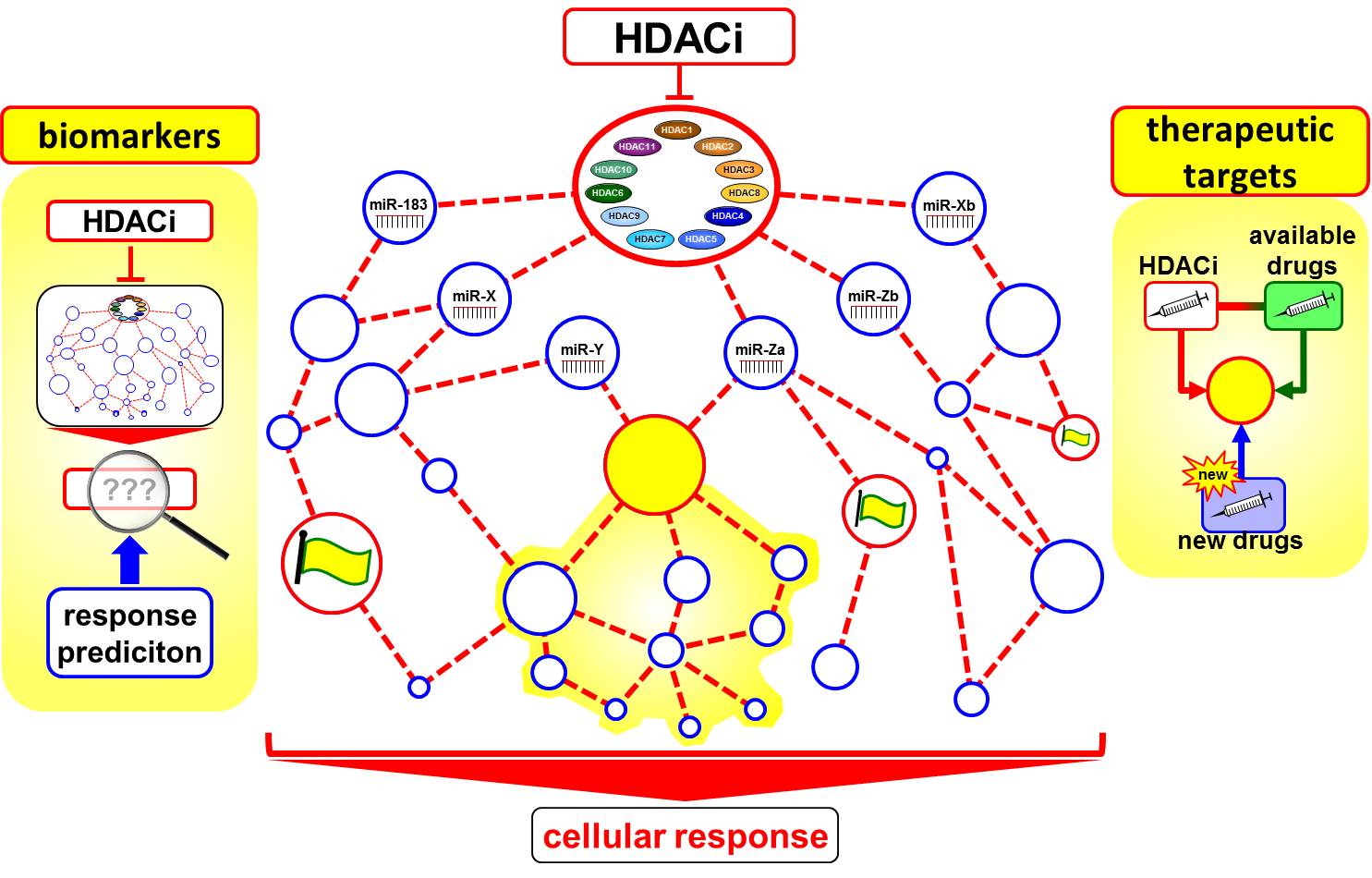
The BE(2)-C neuroblastoma cell line was treated alone or in combination with a BET-bromodomain inhibitor, which indirectly inhibits MYCN activity, and a pan-HDAC inhibitor. A series of concentrations and incubation times were applied before assessing microRNA and mRNA expression using miRNA arrays as data endpoints. The targets of each differentially regulated microRNA will bioinformatically predicted using sequence-based target prediction and integration of mRNA sequencing data. Predicted protein-coding mRNA targets will be then systematically mapped to cellular pathways from the KEGG database and the mirOB database for cancer-specific pathways involving microRNAs among other database resources. The resulting network will be used to address the complexity of epigenetic drug action and related microRNA regulation in neuroblastoma cells by systems biology modeling to identify biomarker for response prediction to HDACi and identification of drug targets for targeted combinatorial treatment approaches in high-risk NB patients.
Keywords: neuroblastoma, systems biology, epigenetics, Bromodomains, Histondeacetylase



